Chapter 10 Tidying Data
Even before we conduct analyses or calculations, we need to put our data into the correct format. The goal here is to rearrange a messy dataset into one that is tidy.
The two most important properties of tidy data are:
- Each column is a variable.
- Each row is an observation.
Tidy data is easier to work with because you have a consistent way of referring to variables (as column names) and observations (as row indices). The data then becomes easier to manipulate, visualize, and model.
For more on the concept of tidy data, read Hadley Wickham’s paper here.
10.1 Wide vs. Long Formats
Tidy datasets are all alike, but every messy dataset is messy in its own way.
Hadley Wickham
Tabular datasets can be arranged in many ways. For instance, consider the data below. Each dataset displays information on heart rates observed in individuals across three different time periods, but the data are organized differently in each table.
wide <- data.frame(
name = c("Wilbur", "Petunia", "Gregory"),
time1 = c(67, 80, 64),
time2 = c(56, 90, 50),
time3 = c(70, 67, 101)
)
wide
#> name time1 time2 time3
#> 1 Wilbur 67 56 70
#> 2 Petunia 80 90 67
#> 3 Gregory 64 50 101
long <- data.frame(
name = c("Wilbur", "Petunia", "Gregory", "Wilbur", "Petunia", "Gregory", "Wilbur", "Petunia", "Gregory"),
time = c(1, 1, 1, 2, 2, 2, 3, 3, 3),
heartrate = c(67, 80, 64, 56, 90, 50, 70, 67, 10)
)
long
#> name time heartrate
#> 1 Wilbur 1 67
#> 2 Petunia 1 80
#> 3 Gregory 1 64
#> 4 Wilbur 2 56
#> 5 Petunia 2 90
#> 6 Gregory 2 50
#> 7 Wilbur 3 70
#> 8 Petunia 3 67
#> 9 Gregory 3 10Question: Which one of these do you think is the tidy format?
Answer: The first dataframe (the “wide” one) would not be considered tidy because values (i.e., heart rate) are spread across multiple columns.
We often refer to these different structures as “long” vs. “wide” formats:
In the “long” format, you usually have one column for the observed variable, and the other columns are ID variables.
In the “wide” format, each row is often a site/subject/patient, and you have multiple observation variables containing the same type of data. These can be either repeated observations over time or observations of multiple variables (or a mix of both). In the case above, we had the same kind of data (heart rate) entered across three different columns, corresponding to three different time periods.
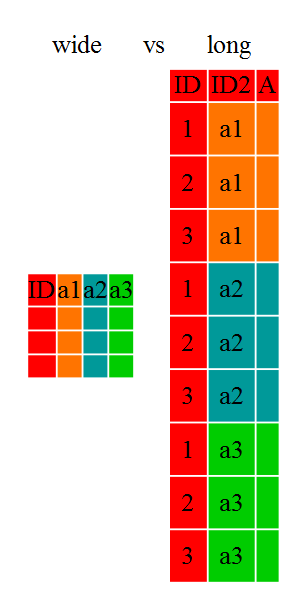
You may find data input in the “wide” format to be simpler, and some other applications may prefer “wide”-format data. However, many of R’s functions have been designed assuming you have “long”-format data.
10.2 Tidying the Gapminder Data
Let’s look at the structure of our original gapminder dataframe:
library(gapminder)
gap <- gapminder
head(gap)
#> # A tibble: 6 x 6
#> country continent year lifeExp pop gdpPercap
#> <fct> <fct> <int> <dbl> <int> <dbl>
#> 1 Afghanistan Asia 1952 28.8 8425333 779.
#> 2 Afghanistan Asia 1957 30.3 9240934 821.
#> 3 Afghanistan Asia 1962 32.0 10267083 853.
#> 4 Afghanistan Asia 1967 34.0 11537966 836.
#> 5 Afghanistan Asia 1972 36.1 13079460 740.
#> 6 Afghanistan Asia 1977 38.4 14880372 786.Question: Is this dataframe wide or long?
Answer: This dataframe is somewhere in between the purely ‘long’ and ‘wide’ formats. We have three “ID variables” (continent, country, year) and three “observation variables” (pop, lifeExp, gdpPercap).
Despite not having all observations in one column, this intermediate format makes sense given that all three observation variables have different units. As we have seen, many of the functions in R are often vector-based, and you usually do not want to do mathematical operations on values with different units.
On the other hand, there are some instances in which a purely long or wide format is ideal (e.g., plotting). Likewise, sometimes you will get data on your desk that is poorly organized, and you will need to reshape it.
10.3 tidyr Functions
Thankfully, the tidyr package will help you efficiently transform your data regardless of their original format.
10.3.1 gather
Until now, we have been using the nicely formatted original gapminder dataset. This dataset is not quite wide and not quite long – it is something in the middle – but ‘real’ data (i.e., our own research data) will never be so well organized. Here let’s start with the wide-format version of the gapminder dataset.
gap_wide <- read.csv("data/gapminder_wide.csv", stringsAsFactors = FALSE)
head(gap_wide)
#> continent country gdpPercap_1952 gdpPercap_1957 gdpPercap_1962
#> 1 Africa Algeria 2449 3014 2551
#> 2 Africa Angola 3521 3828 4269
#> 3 Africa Benin 1063 960 949
#> 4 Africa Botswana 851 918 984
#> 5 Africa Burkina Faso 543 617 723
#> 6 Africa Burundi 339 380 355
#> gdpPercap_1967 gdpPercap_1972 gdpPercap_1977 gdpPercap_1982 gdpPercap_1987
#> 1 3247 4183 4910 5745 5681
#> 2 5523 5473 3009 2757 2430
#> 3 1036 1086 1029 1278 1226
#> 4 1215 2264 3215 4551 6206
#> 5 795 855 743 807 912
#> 6 413 464 556 560 622
#> gdpPercap_1992 gdpPercap_1997 gdpPercap_2002 gdpPercap_2007 lifeExp_1952
#> 1 5023 4797 5288 6223 43.1
#> 2 2628 2277 2773 4797 30.0
#> 3 1191 1233 1373 1441 38.2
#> 4 7954 8647 11004 12570 47.6
#> 5 932 946 1038 1217 32.0
#> 6 632 463 446 430 39.0
#> lifeExp_1957 lifeExp_1962 lifeExp_1967 lifeExp_1972 lifeExp_1977 lifeExp_1982
#> 1 45.7 48.3 51.4 54.5 58.0 61.4
#> 2 32.0 34.0 36.0 37.9 39.5 39.9
#> 3 40.4 42.6 44.9 47.0 49.2 50.9
#> 4 49.6 51.5 53.3 56.0 59.3 61.5
#> 5 34.9 37.8 40.7 43.6 46.1 48.1
#> 6 40.5 42.0 43.5 44.1 45.9 47.5
#> lifeExp_1987 lifeExp_1992 lifeExp_1997 lifeExp_2002 lifeExp_2007 pop_1952
#> 1 65.8 67.7 69.2 71.0 72.3 9279525
#> 2 39.9 40.6 41.0 41.0 42.7 4232095
#> 3 52.3 53.9 54.8 54.4 56.7 1738315
#> 4 63.6 62.7 52.6 46.6 50.7 442308
#> 5 49.6 50.3 50.3 50.6 52.3 4469979
#> 6 48.2 44.7 45.3 47.4 49.6 2445618
#> pop_1957 pop_1962 pop_1967 pop_1972 pop_1977 pop_1982 pop_1987 pop_1992
#> 1 10270856 11000948 12760499 14760787 17152804 20033753 23254956 26298373
#> 2 4561361 4826015 5247469 5894858 6162675 7016384 7874230 8735988
#> 3 1925173 2151895 2427334 2761407 3168267 3641603 4243788 4981671
#> 4 474639 512764 553541 619351 781472 970347 1151184 1342614
#> 5 4713416 4919632 5127935 5433886 5889574 6634596 7586551 8878303
#> 6 2667518 2961915 3330989 3529983 3834415 4580410 5126023 5809236
#> pop_1997 pop_2002 pop_2007
#> 1 29072015 31287142 33333216
#> 2 9875024 10866106 12420476
#> 3 6066080 7026113 8078314
#> 4 1536536 1630347 1639131
#> 5 10352843 12251209 14326203
#> 6 6121610 7021078 8390505The first step towards getting our nice intermediate data format is to first convert from the wide to the long format.
The function gather() will ‘gather’ the observation variables into a single variable. This is sometimes called “melting” your data, because it melts the table from wide to long. Those data will be melted into two variables: one for the variable names and the other for the variable values.
gap_long <- gap_wide %>%
gather(obstype_year, obs_values, 3:38)
head(gap_long)
#> continent country obstype_year obs_values
#> 1 Africa Algeria gdpPercap_1952 2449
#> 2 Africa Angola gdpPercap_1952 3521
#> 3 Africa Benin gdpPercap_1952 1063
#> 4 Africa Botswana gdpPercap_1952 851
#> 5 Africa Burkina Faso gdpPercap_1952 543
#> 6 Africa Burundi gdpPercap_1952 339Notice that we put three arguments into the gather() function:
- The name for the new ID variable (
obstype_year). - The name for the new amalgamated observation variable (
obs_value). - The indices of the old observation variables (
3:38, signalling columns 3 through 38) that we want to gather into one variable. Notice that we do not want to melt down columns 1 and 2, as these are considered ID variables.
We can select observation variables using:
- Variable indices.
- Variable names (without quotes).
x:zto select all variables between x and z.-yto exclude y.starts_with(x, ignore.case = TRUE): All names that start withx.ends_with(x, ignore.case = TRUE): All names that end withx.contains(x, ignore.case = TRUE): All names that containx.
See the select() function in dplyr for more options.
For instance, here we do the same thing with (1) the starts_with function and (2) the - operator:
# 1. With the starts_with() function:
gap_long <- gap_wide %>%
gather(obstype_year, obs_values, starts_with('pop'),
starts_with('lifeExp'), starts_with('gdpPercap'))
head(gap_long)
#> continent country obstype_year obs_values
#> 1 Africa Algeria pop_1952 9279525
#> 2 Africa Angola pop_1952 4232095
#> 3 Africa Benin pop_1952 1738315
#> 4 Africa Botswana pop_1952 442308
#> 5 Africa Burkina Faso pop_1952 4469979
#> 6 Africa Burundi pop_1952 2445618
# 2. With the - operator:
gap_long <- gap_wide %>%
gather(obstype_year, obs_values, -continent, -country)
head(gap_long)
#> continent country obstype_year obs_values
#> 1 Africa Algeria gdpPercap_1952 2449
#> 2 Africa Angola gdpPercap_1952 3521
#> 3 Africa Benin gdpPercap_1952 1063
#> 4 Africa Botswana gdpPercap_1952 851
#> 5 Africa Burkina Faso gdpPercap_1952 543
#> 6 Africa Burundi gdpPercap_1952 339However you choose to do it, notice that the output collapses all of the measured variables into two columns: one containing the new ID variable, the other containing the observation value for that row.
10.3.2 separate
You will notice that, in our long dataset, obstype_year actually contains two pieces of information, the observation type (pop, lifeExp, or gdpPercap) and the year.
We can use the separate() function to split the character strings into multiple variables.
gap_long_sep <- gap_long %>%
separate(obstype_year, into = c('obs_type','year'), sep = "_") %>%
mutate(year = as.integer(year))
head(gap_long_sep)
#> continent country obs_type year obs_values
#> 1 Africa Algeria gdpPercap 1952 2449
#> 2 Africa Angola gdpPercap 1952 3521
#> 3 Africa Benin gdpPercap 1952 1063
#> 4 Africa Botswana gdpPercap 1952 851
#> 5 Africa Burkina Faso gdpPercap 1952 543
#> 6 Africa Burundi gdpPercap 1952 33910.3.3 spread
The opposite of gather() is spread(). It spreads our observation variables back out to make a wider table. We can use this function to spread our gap_long() to the original “medium” format.
gap_medium <- gap_long_sep %>%
spread(obs_type, obs_values)
head(gap_medium)
#> continent country year gdpPercap lifeExp pop
#> 1 Africa Algeria 1952 2449 43.1 9279525
#> 2 Africa Algeria 1957 3014 45.7 10270856
#> 3 Africa Algeria 1962 2551 48.3 11000948
#> 4 Africa Algeria 1967 3247 51.4 12760499
#> 5 Africa Algeria 1972 4183 54.5 14760787
#> 6 Africa Algeria 1977 4910 58.0 17152804All we need is some quick fixes to make this dataset identical to the original gapminder dataset:
gap <- gapminder
head(gap)
#> # A tibble: 6 x 6
#> country continent year lifeExp pop gdpPercap
#> <fct> <fct> <int> <dbl> <int> <dbl>
#> 1 Afghanistan Asia 1952 28.8 8425333 779.
#> 2 Afghanistan Asia 1957 30.3 9240934 821.
#> 3 Afghanistan Asia 1962 32.0 10267083 853.
#> 4 Afghanistan Asia 1967 34.0 11537966 836.
#> 5 Afghanistan Asia 1972 36.1 13079460 740.
#> 6 Afghanistan Asia 1977 38.4 14880372 786.
# Rearrange columns:
gap_medium <- gap_medium %>%
select(country, continent, year, lifeExp, pop, gdpPercap)
head(gap_medium)
#> country continent year lifeExp pop gdpPercap
#> 1 Algeria Africa 1952 43.1 9279525 2449
#> 2 Algeria Africa 1957 45.7 10270856 3014
#> 3 Algeria Africa 1962 48.3 11000948 2551
#> 4 Algeria Africa 1967 51.4 12760499 3247
#> 5 Algeria Africa 1972 54.5 14760787 4183
#> 6 Algeria Africa 1977 58.0 17152804 4910
# Arrange by country, continent, and year:
gap_medium <- gap_medium %>%
arrange(country,continent,year)
head(gap_medium)
#> country continent year lifeExp pop gdpPercap
#> 1 Afghanistan Asia 1952 28.8 8425333 779
#> 2 Afghanistan Asia 1957 30.3 9240934 821
#> 3 Afghanistan Asia 1962 32.0 10267083 853
#> 4 Afghanistan Asia 1967 34.0 11537966 836
#> 5 Afghanistan Asia 1972 36.1 13079460 740
#> 6 Afghanistan Asia 1977 38.4 14880372 786What we just told you will become obsolete…
gather and spread are being replaced by pivot_longer and pivot_wider in tidyr 1.0.0, which uses ideas from the cdata package to make reshaping easier to think about. In future classes, we will migrate to those functions.
10.4 Dealing with Missing Data
A common challenge in applying quantitative tools to social science problems is dealing with missing data. You’ll see a variety of ways the creators of data sets designate that a piece of data is missing - for example, NA, -99, or -77 are sometimes used to denote a missing piece of data. We recommend using NA, which has a variety of associated functions that are useful when transforming missing data.
10.4.1 na_if
You can use na_if to replace certain pieces of data with NA. Consider the case where lifeExp values below 35 are missing and simply filled in with “unknown”:
This is problematic for many reasons, including that we cannot perform simple mathematical functions on columns with both number and character values:
mean(gap_medium$lifeExp, na.rm = TRUE)
#> Warning in mean.default(gap_medium$lifeExp, na.rm = TRUE): argument is not
#> numeric or logical: returning NA
#> [1] NAHowever, NAs are different in the sense that they can exist in a numeric vector, and therefore you still can perform math functions (R will omit those observations with NA). Below, we replace the “unknown” values with NA, and we can then calculate the mean lifeExp.
10.4.2 Replace NA values with replace_na
Sometimes, you will want to replace all NA values in your data (for instance, maybe you know that the true value of anything coded as NA is actually 30). replace_na is a simple command that will replace all NAs with a new value.
gap_na_replaced <- gap_medium %>%
mutate(lifeExp = replace_na(lifeExp, 30))
head(gap_na_replaced)
#> country continent year lifeExp pop gdpPercap
#> 1 Afghanistan Asia 1952 30.0 8425333 779
#> 2 Afghanistan Asia 1957 30.0 9240934 821
#> 3 Afghanistan Asia 1962 30.0 10267083 853
#> 4 Afghanistan Asia 1967 30.0 11537966 836
#> 5 Afghanistan Asia 1972 36.1 13079460 740
#> 6 Afghanistan Asia 1977 38.4 14880372 78610.5 More tidyverse
dplyr and tidyr have many more functions to help you wrangle and manipulate
your data. See the Data Wrangling Cheatsheet for more.
There are some other useful packages in the tidyverse:
ggplot2for plotting (we will cover this in the Visualization module).readrandhavenfor reading in data.purrrfor working iterations.stringr,lubridate, andforcatsfor manipulating strings, dates, and factors, respectively.- Many many more! Take a peak at the tidyverse GitHub page…
10.6 Challenges
Challenge 1.
Subset the results from Challenge #3 (of the previous chapter) to select only the country, year, and gdpPercap_diff columns. Use tidyr to put it in wide format so that countries are rows and years are columns.
Challenge 2.
Now turn the dataframe above back into the long format with three columns: country, year, and gdpPercap_diff.
Acknowledgments
Some of the materials in this module were adapted from: